|
chrX_+_12993224
|
0.778
|
NM_021109
|
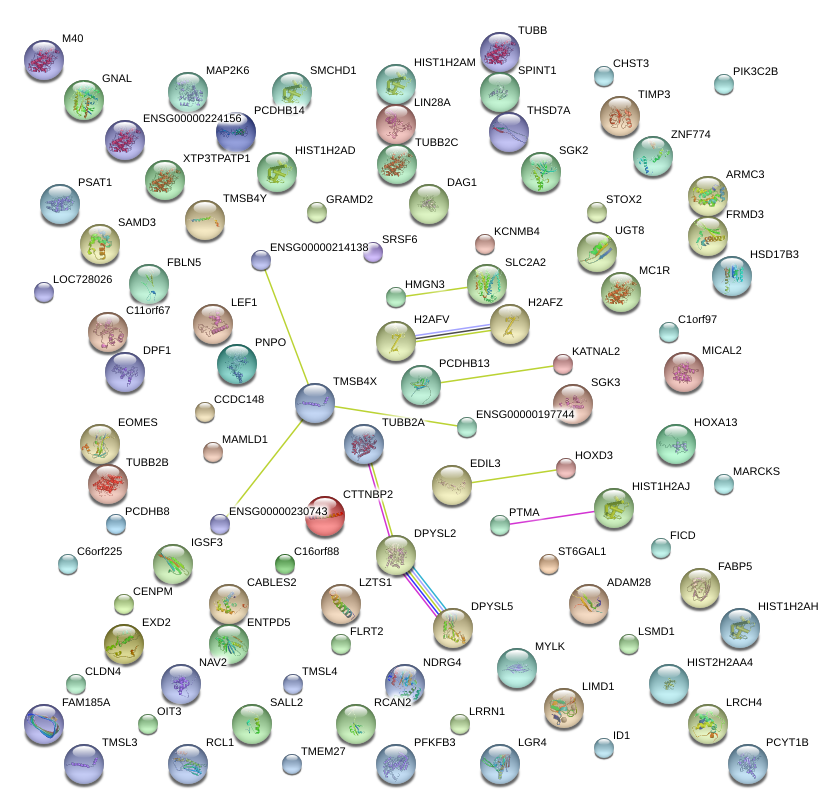
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr6_+_27100784
|
0.711
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr1_+_26737306
|
0.623
|
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr15_+_90895476
|
0.602
|
NM_001004309
|
ZNF774
|
zinc finger protein 774
|
|
chr11_+_77532159
|
0.562
|
NM_024684
|
C11orf67
|
chromosome 11 open reading frame 67
|
|
chr8_+_82192717
|
0.552
|
NM_001444
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated)
|
|
chrX_-_24690740
|
0.528
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr8_+_24151579
|
0.517
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr7_-_117513466
|
0.474
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr11_+_10830752
|
0.440
|
|
|
|
|
chr14_-_90421032
|
0.432
|
NM_145231
|
EFCAB11
|
EF-hand calcium binding domain 11
|
|
chr6_+_27114860
|
0.416
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr6_+_30688085
|
0.396
|
|
TUBB
|
tubulin, beta class I
|
|
chr12_+_70759974
|
0.393
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr6_+_30688144
|
0.385
|
NM_178014
|
TUBB
|
tubulin, beta class I
|
|
chr15_+_41136622
|
0.379
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr5_-_83680191
|
0.372
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr14_+_85996512
|
0.369
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr8_+_26435339
|
0.368
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chrX_-_15683153
|
0.365
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr1_-_117210301
|
0.363
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr20_+_30193083
|
0.345
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr4_+_184827805
|
0.343
|
|
STOX2
|
storkhead box 2
|
|
chr17_+_46019022
|
0.342
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr11_+_20049128
|
0.342
|
|
NAV2
|
neuron navigator 2
|
|
chr8_-_20112691
|
0.339
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr2_+_177028804
|
0.338
|
NM_006898
|
HOXD3
|
homeobox D3
|
|
chr3_-_123339178
|
0.337
|
|
MYLK
|
myosin light chain kinase
|
|
chr4_+_184827882
|
0.334
|
|
|
|
|
chr22_+_33197678
|
0.326
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr8_+_26435590
|
0.325
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_+_140602914
|
0.324
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr9_-_99064385
|
0.323
|
NM_000197
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3
|
|
chr7_+_73245192
|
0.323
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr3_-_123339423
|
0.318
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr3_+_186648515
|
0.318
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr7_-_44887567
|
0.315
|
|
H2AFV
|
H2A histone family, member V
|
|
chr22_+_33197744
|
0.314
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr12_+_108909049
|
0.314
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr6_-_79944397
|
0.308
|
NM_001201362
NM_001201363
NM_004242
NM_138730
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr17_+_67498391
|
0.306
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr6_+_114178516
|
0.302
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr1_-_204463512
|
0.301
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr7_-_44887697
|
0.298
|
NM_012412
NM_138635
NM_201436
NM_201516
NM_201517
|
H2AFV
|
H2A histone family, member V
|
|
chr7_-_44887620
|
0.298
|
|
H2AFV
|
H2A histone family, member V
|
|
chr17_+_46018888
|
0.298
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr7_-_27239724
|
0.298
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr18_+_3594056
|
0.297
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr10_+_6244839
|
0.293
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr14_+_85996454
|
0.290
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr9_+_80911990
|
0.288
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr7_-_11871735
|
0.283
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr20_-_60982334
|
0.283
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr14_+_69658193
|
0.282
|
NM_001193360
NM_001193361
NM_018199
|
EXD2
|
exonuclease 3'-5' domain containing 2
|
|
chrX_+_149613719
|
0.280
|
NM_001177466
NM_005491
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr18_-_19748928
|
0.280
|
|
LOC100128893
|
uncharacterized LOC100128893
|
|
chr9_-_85882093
|
0.279
|
NM_001244962
|
FRMD3
|
FERM domain containing 3
|
|
chr4_+_115543522
|
0.278
|
NM_003360
|
UGT8
|
UDP glycosyltransferase 8
|
|
chr6_-_79944324
|
0.274
|
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr7_+_102389325
|
0.273
|
NM_001145268
NM_001145269
|
FAM185A
|
family with sequence similarity 185, member A
|
|
chr3_+_49507470
|
0.271
|
NM_001165928
NM_001177634
NM_001177635
NM_001177636
NM_001177637
NM_001177638
NM_001177639
NM_001177640
NM_001177641
NM_001177642
NM_001177644
NM_004393
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr7_-_44887663
|
0.271
|
|
H2AFV
|
H2A histone family, member V
|
|
chr22_-_42336222
|
0.271
|
NM_001110215
|
CENPM
|
centromere protein M
|
|
chr9_+_4792974
|
0.269
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr4_-_109089444
|
0.265
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chrX_-_73512376
|
0.264
|
|
FTX
|
FTX transcript, XIST regulator (non-protein coding)
|
|
chr5_+_140557370
|
0.264
|
NM_019120
|
PCDHB8
|
protocadherin beta 8
|
|
chr14_-_92413738
|
0.264
|
|
FBLN5
|
fibulin 5
|
|
chr17_-_7760740
|
0.261
|
|
LSMD1
|
LSM domain containing 1
|
|
chr11_-_27494232
|
0.259
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr6_+_112408673
|
0.259
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr3_+_3841079
|
0.259
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr14_+_69658215
|
0.257
|
|
EXD2
|
exonuclease 3'-5' domain containing 2
|
|
chr3_-_170744458
|
0.255
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr4_+_184826427
|
0.252
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr10_+_74653338
|
0.250
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr5_+_140593508
|
0.245
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr18_+_44497512
|
0.245
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr10_+_23216953
|
0.245
|
NM_173081
|
ARMC3
|
armadillo repeat containing 3
|
|
chr18_+_11751470
|
0.243
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr3_+_45636176
|
0.240
|
NM_014240
|
LIMD1
|
LIM domains containing 1
|
|
chr3_-_27764197
|
0.238
|
|
EOMES
|
eomesodermin
|
|
chr20_-_43024303
|
0.237
|
|
|
|
|
chr20_+_42086533
|
0.236
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chrX_-_24665454
|
0.235
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr15_-_72490109
|
0.231
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr14_-_21994342
|
0.230
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr2_+_27070962
|
0.230
|
NM_020134
NM_001253723
NM_001253724
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr2_+_232573226
|
0.230
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr20_+_42194736
|
0.229
|
NM_016276
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr6_-_27782517
|
0.228
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr6_-_46293215
|
0.227
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_-_38720330
|
0.226
|
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr10_+_73723992
|
0.225
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr14_+_69658482
|
0.224
|
NM_001193362
NM_001193363
|
EXD2
|
exonuclease 3'-5' domain containing 2
|
|
chr6_-_130536417
|
0.223
|
NM_152552
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr2_-_159312955
|
0.222
|
NM_001171637
NM_138803
|
CCDC148
|
coiled-coil domain containing 148
|
|
chr2_+_232573249
|
0.220
|
|
PTMA
|
prothymosin, alpha
|
|
chr16_+_58534045
|
0.220
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr11_+_12132063
|
0.220
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr1_+_151032906
|
0.219
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr19_-_8455552
|
0.219
|
|
LOC100507567
|
uncharacterized LOC100507567
|
|
chr6_+_41040713
|
0.219
|
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chrX_-_112084006
|
0.217
|
NM_133265
|
AMOT
|
angiomotin
|
|
chrX_+_107288199
|
0.216
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr5_-_151150939
|
0.215
|
|
LOC100652758
|
uncharacterized LOC100652758
|
|
chrX_-_133931146
|
0.214
|
NM_001166599
NM_001166600
NM_001170757
|
FAM122B
|
family with sequence similarity 122B
|
|
chr7_+_139528951
|
0.212
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr1_+_64239566
|
0.210
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr15_+_41136178
|
0.210
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr17_-_38821286
|
0.209
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr5_+_140729723
|
0.207
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr1_+_10459084
|
0.206
|
NM_002631
|
PGD
|
phosphogluconate dehydrogenase
|
|
chr16_-_46782104
|
0.206
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr10_-_13390189
|
0.205
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr1_+_213123853
|
0.203
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr5_+_40680021
|
0.203
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr1_+_164528914
|
0.201
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr16_+_15528324
|
0.201
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr2_-_45236521
|
0.200
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr16_-_71610948
|
0.199
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr1_+_151032866
|
0.198
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chrX_+_18443702
|
0.197
|
NM_003159
|
CDKL5
|
cyclin-dependent kinase-like 5
|
|
chr7_-_27205148
|
0.196
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr3_+_181429711
|
0.195
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr17_-_10372875
|
0.195
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr15_-_34659245
|
0.195
|
NM_153613
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4
|
|
chr22_+_24115035
|
0.194
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr16_-_838374
|
0.194
|
NM_058192
|
RPUSD1
|
RNA pseudouridylate synthase domain containing 1
|
|
chr3_+_45635353
|
0.193
|
|
LIMD1
|
LIM domains containing 1
|
|
chr5_-_146833150
|
0.192
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr1_+_151032880
|
0.192
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr4_+_41614911
|
0.191
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_-_167883243
|
0.191
|
NM_001167749
NM_018417
|
ADCY10
|
adenylate cyclase 10 (soluble)
|
|
chr10_-_103599596
|
0.191
|
NM_173194
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr10_+_115310589
|
0.190
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr3_-_14989857
|
0.190
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr14_+_103800534
|
0.190
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr5_+_140739589
|
0.190
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr10_+_24498088
|
0.190
|
|
KIAA1217
|
KIAA1217
|
|
chr9_+_127420690
|
0.190
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr5_+_140787769
|
0.189
|
NM_018926
NM_032100
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr7_-_83270746
|
0.188
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chrX_-_55187561
|
0.188
|
NM_001166702
NM_001166699
NM_001166700
NM_001166701
NM_001166704
NM_138362
|
FAM104B
|
family with sequence similarity 104, member B
|
|
chr1_+_174843523
|
0.186
|
NM_001243763
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr17_+_26646120
|
0.185
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr14_-_65409445
|
0.185
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr18_-_74728677
|
0.184
|
|
MBP
|
myelin basic protein
|
|
chr11_-_71814268
|
0.183
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr12_-_70093064
|
0.183
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr6_-_26043884
|
0.182
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr17_-_39684585
|
0.182
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr2_+_189839132
|
0.181
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr22_+_29196668
|
0.181
|
|
|
|
|
chr2_-_100722328
|
0.181
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr5_-_41510627
|
0.179
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr17_+_68071386
|
0.178
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr17_-_56595192
|
0.177
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr1_-_6479880
|
0.176
|
NM_019089
|
HES2
|
hairy and enhancer of split 2 (Drosophila)
|
|
chr17_-_39684499
|
0.175
|
|
KRT19
|
keratin 19
|
|
chr6_-_13487772
|
0.175
|
NM_001242629
NM_018988
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr3_-_185826748
|
0.174
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr19_-_45004482
|
0.174
|
NM_013256
|
ZNF180
|
zinc finger protein 180
|
|
chr20_+_42086468
|
0.174
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr1_-_26324534
|
0.172
|
NM_000437
|
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa
|
|
chr14_+_50087400
|
0.172
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr15_-_59041933
|
0.172
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr15_-_59041812
|
0.171
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr15_+_39873276
|
0.171
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr7_-_41742666
|
0.170
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr2_+_74881353
|
0.169
|
NM_004263
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F
|
|
chr4_-_40936640
|
0.169
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr5_-_111093251
|
0.168
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_75915566
|
0.167
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr8_-_125580745
|
0.167
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr10_+_24497719
|
0.166
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr1_+_213124008
|
0.165
|
|
VASH2
|
vasohibin 2
|
|
chr7_-_107443631
|
0.165
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr2_-_128421965
|
0.165
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr16_-_21436657
|
0.165
|
NM_130464
|
NPIPL3
|
nuclear pore complex interacting protein-like 3
|
|
chr1_+_50571963
|
0.165
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr15_-_64673569
|
0.164
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr18_-_21891469
|
0.163
|
NM_001242508
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr1_-_32801606
|
0.163
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr2_-_175869910
|
0.161
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr10_-_14646237
|
0.160
|
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr6_+_41040891
|
0.160
|
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chr12_-_81763127
|
0.160
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr1_-_16539103
|
0.159
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr1_+_151032683
|
0.159
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr18_-_24128499
|
0.158
|
NM_001142730
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr7_+_96635289
|
0.158
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr19_+_7694658
|
0.158
|
NM_001171155
|
C19orf79
|
chromosome 19 open reading frame 79
|
|
chr2_+_223536362
|
0.158
|
NM_058165
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1
|
|
chr1_+_24834083
|
0.157
|
NM_001251978
|
RCAN3
|
RCAN family member 3
|
|
chr6_+_105404922
|
0.157
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr7_-_92855780
|
0.157
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr3_-_149688886
|
0.157
|
|
PFN2
|
profilin 2
|
|
chr12_-_56694123
|
0.157
|
|
CS
|
citrate synthase
|
|
chr1_+_87797301
|
0.157
|
|
|
|